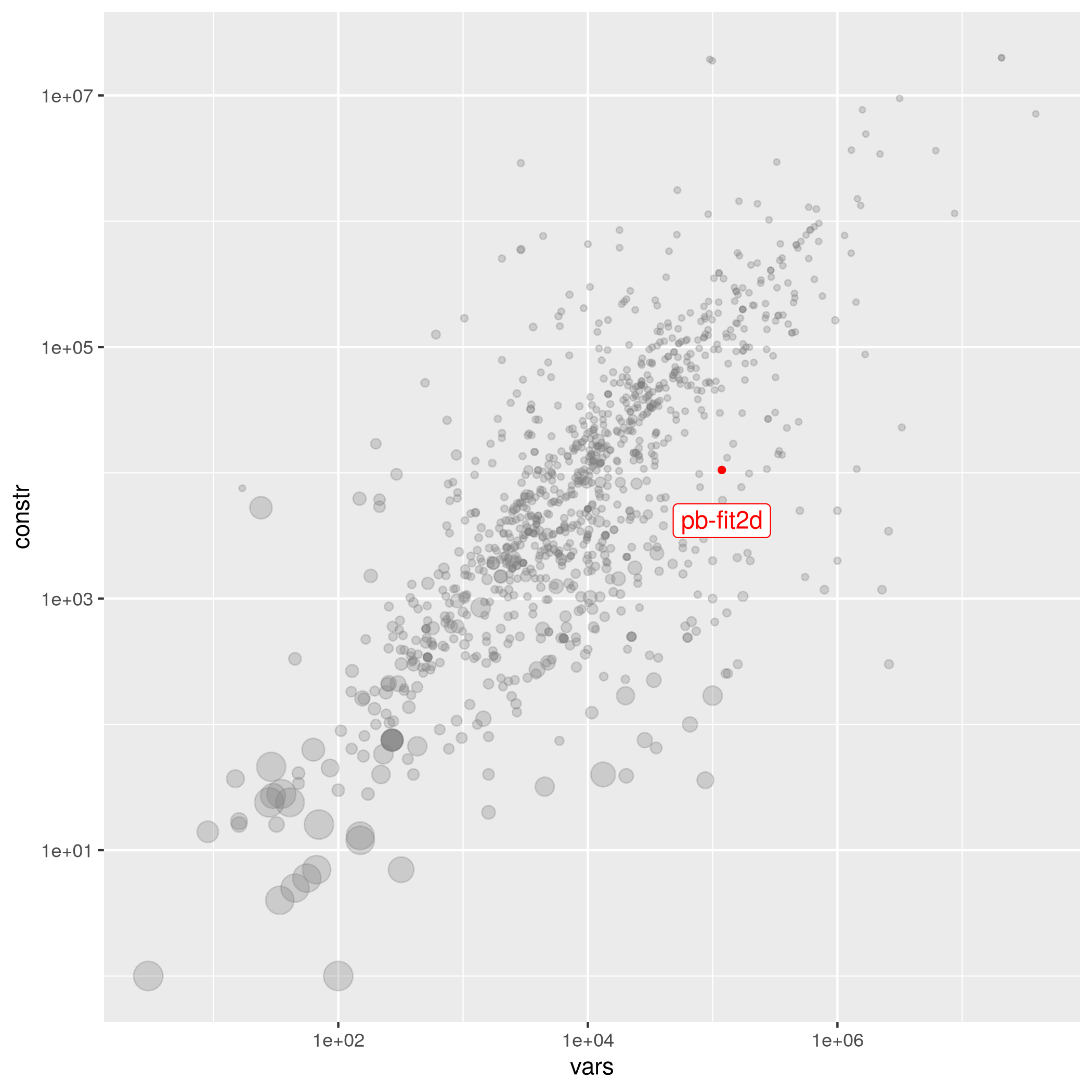
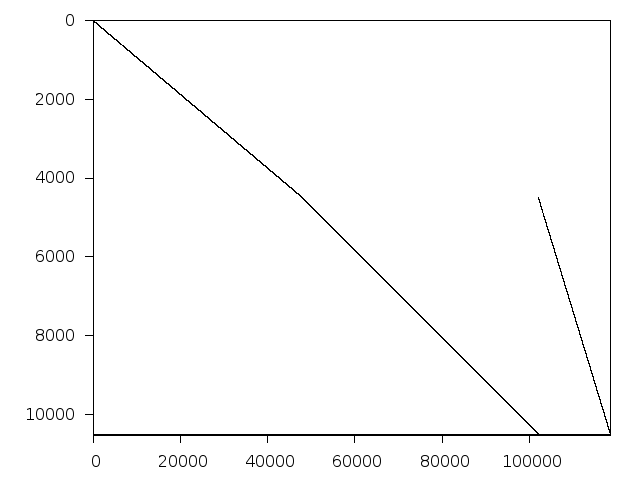
pb-fit2d
binary decomposition benchmark_suitable equation_knapsack binpacking knapsack
| Submitter | Variables | Constraints | Density | Status | Group | Objective | MPS File |
|---|---|---|---|---|---|---|---|
| Gleb Belov | 118500 | 10525 | 1.26328e-03 | easy | pb- | -20425 | pb-fit2d.mps.gz |
These are the instances from MiniZinc Challenges 2012-2016 (see www.minizinc.org), compiled for MIP WITH INDICATOR CONSTRAINTS using the develop branch of MiniZinc and CPLEX 12.7.1 on 30 April 2017. Thus, these instances can only be handled by solvers accepting indicator constraints. For instances compiled with big-M/domain decomposition only, see my previous submission to MIPLIB. To recompile, create a directory MODELS, a list lst12_16.txt of the instances with full paths to mzn/dzn files of each instance per line, and say $> ~/install/libmzn/tests/benchmarking/mzn-test.py -l ../lst12_16.txt –slvPrf MZN-CPLEX –debug 1 –addOption “–timeout 3 -D fIndConstr=true -D fMIPdomains=false” –useJoinedName “–writeModel MODELS_IND/%s.mps” Alternatively, you can compile individual instance as follows: $> mzn-cplex -v -s -G linear –output-time ../challenge_2012_2016/mznc2016_probs/zephyrus/zephyrus.mzn ../challenge_2012_2016/mznc2016_p/zephyrus/14__8__6__3.dzn -a –timeout 3 -D fIndConstr=true -D fMIPdomains=false –writeModel MODELS_IND/challenge_2012_2016mznc2016_probszephyruszephyrusmzn-challenge_2012_2016mznc2016_probszephyrus14__8__6__3dzn.mps Imported from the MIPLIB2010 submissions.
Instance Statistics
Detailed explanation of the following tables can be found here.
| Original | Presolved | |
|---|---|---|
| Variables | 118500 | 118500 |
| Constraints | 10525 | 10525 |
| Binaries | 118500 | 118500 |
| Integers | 0 | 0 |
| Continuous | 0 | 0 |
| Implicit Integers | 0 | 0 |
| Fixed Variables | 0 | 0 |
| Nonzero Density | 0.00126328 | 0.00126328 |
| Nonzeroes | 1575580 | 1575580 |
| Original | Presolved | |
|---|---|---|
| Total | 10525 | 10525 |
| Empty | 0 | 0 |
| Free | 0 | 0 |
| Singleton | 0 | 0 |
| Aggregations | 0 | 0 |
| Precedence | 0 | 0 |
| Variable Bound | 0 | 0 |
| Set Partitioning | 0 | 0 |
| Set Packing | 0 | 0 |
| Set Covering | 0 | 0 |
| Cardinality | 0 | 0 |
| Invariant Knapsack | 0 | 0 |
| Equation Knapsack | 1 | 1 |
| Bin Packing | 0 | 10500 |
| Knapsack | 0 | 24 |
| Integer Knapsack | 0 | 0 |
| Mixed Binary | 10524 | 0 |
| General Linear | 0 | 0 |
| Indicator | 0 | 0 |
Structure
Available nonzero structure and decomposition information. Further information can be found here.

Decomposed structure of original problem (dec-file)

Decomposed structure after trivial presolving (dec-file)
| value | min | median | mean | max | |
|---|---|---|---|---|---|
| Components | 4.021231 | ||||
| Constraint % | 0.00950 | 0.00950 | 0.00950 | 0.0095000 | |
| Variable % | 0.00844 | 0.00952 | 0.00928 | 0.0118143 | |
| Score | 0.997530 |
Best Known Solution(s)
Find solutions below. Download the archive containing all solutions from the Download page.
| ID | Objective | Exact | Int. Viol | Cons. Viol | Obj. Viol | Submitter | Date | Description |
|---|---|---|---|---|---|---|---|---|
| 1 | -20425 | -20425 | 0 | 0 | 0 | - | 2018-10-11 | Solution found during MIPLIB2017 problem selection. |
Similar instances in collection
The following instances are most similar to pb-fit2d in the collection. This similarity analysis is based on 100 scaled instance features describing properties of the variables, objective function, bounds, constraints, and right hand sides.
| Instance | Status | Variables | Binaries | Integers | Continuous | Constraints | Nonz. | Submitter | Group | Objective | Tags |
|---|---|---|---|---|---|---|---|---|---|---|---|
| neos-787933 | easy | 236376 | 236376 | 0 | 0 | 1897 | 298320 | NEOS Server Submission | neos-pseudoapplication-36 | 30 | benchmark binary decomposition benchmark_suitable binpacking mixed_binary |
| co-100 | easy | 48417 | 48417 | 0 | 0 | 2187 | 1995817 | Axel Werner | – | 2639942.06 | benchmark binary benchmark_suitable precedence set_partitioning set_packing binpacking knapsack |
| neos-2328163-agri | easy | 2236 | 2236 | 0 | 0 | 1963 | 12740 | Jeff Linderoth | neos-pseudoapplication-36 | 27674 | binary decomposition benchmark_suitable set_partitioning set_packing set_covering cardinality invariant_knapsack binpacking knapsack |
| peg-solitaire-a3 | easy | 4552 | 4552 | 0 | 0 | 4587 | 28387 | Hiroshige Dan ; Koichi Fujii | pegsolitaire | 1 | benchmark binary benchmark_suitable aggregations variable_bound set_partitioning cardinality binpacking |
| neos-4355351-swalm | open | 21065 | 10530 | 0 | 10535 | 21609 | 371467 | Jeff Linderoth | neos-pseudoapplication-58 | 33.45757454008309* | variable_bound binpacking mixed_binary |
Reference
No bibliographic information availableLast Update 2024 by Julian Manns
generated with R Markdown
© by Zuse Institute Berlin (ZIB)
Imprint