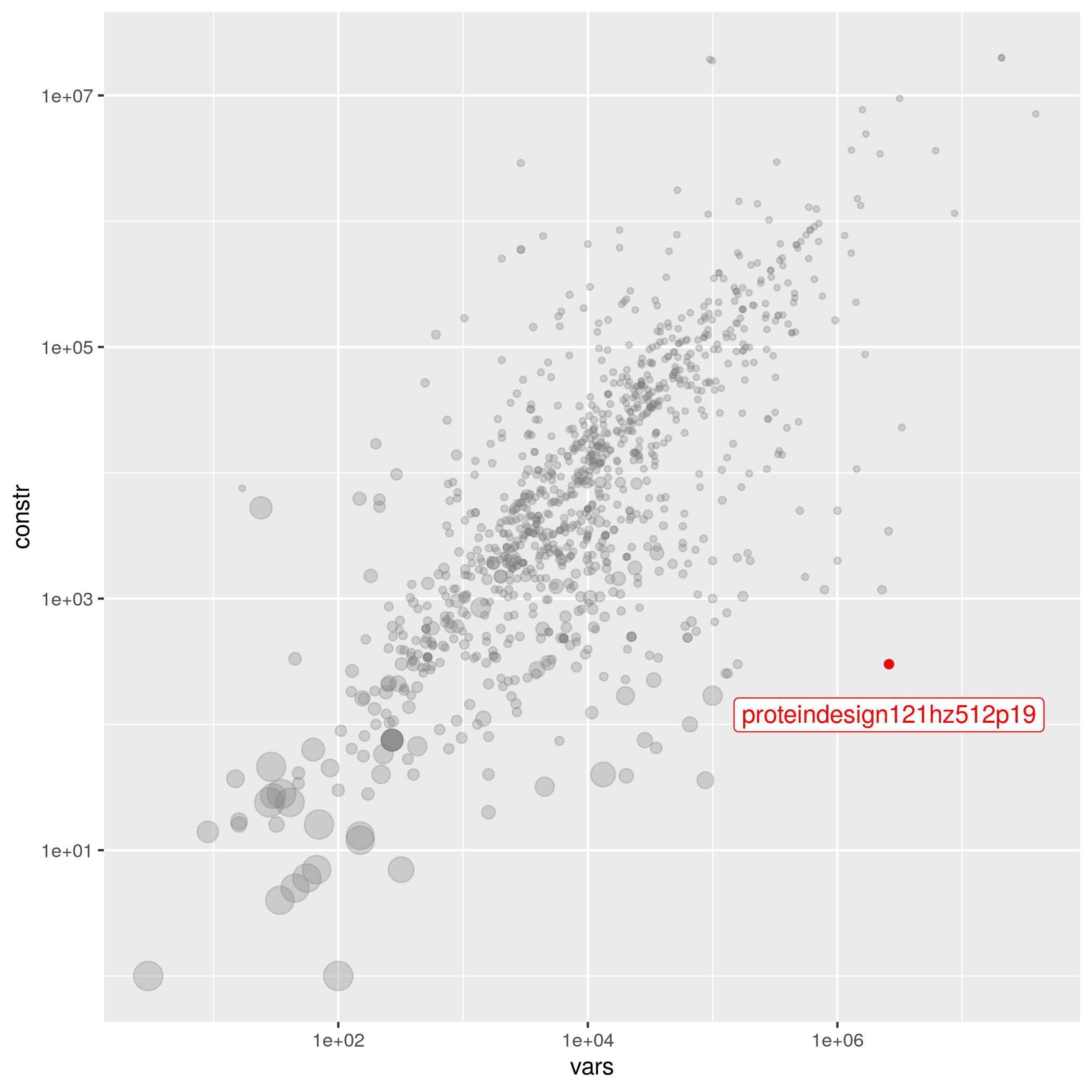
proteindesign121hz512p19
numerics set_partitioning general_linear
| Submitter | Variables | Constraints | Density | Status | Group | Objective | MPS File |
|---|---|---|---|---|---|---|---|
| Gleb Belov | 2589931 | 301 | 1.32524e-02 | open | proteindesign | 3382.0* | proteindesign121hz512p19.mps.gz |
Linearized Constraint Programming models of the MiniZinc Challenges 2012-2016. I should be able to produce versions with indicator constraints supported by Gurobi and CPLEX, however don’t know if you can use them and if there is a standard format. These MPS were produced by Gurobi 7.0.2 using the MiniZinc develop branch on eb536656062ca13325a96b5d0881742c7d0e3c38
Instance Statistics
Detailed explanation of the following tables can be found here.
| Original | Presolved | |
|---|---|---|
| Variables | 2589931 | 2589931 |
| Constraints | 301 | 301 |
| Binaries | 2589840 | 2589840 |
| Integers | 91 | 91 |
| Continuous | 0 | 0 |
| Implicit Integers | 0 | 12 |
| Fixed Variables | 0 | 0 |
| Nonzero Density | 0.0132524 | 0.0132524 |
| Nonzeroes | 10331100 | 10331100 |
| Original | Presolved | |
|---|---|---|
| Total | 301 | 301 |
| Empty | 0 | 0 |
| Free | 0 | 0 |
| Singleton | 0 | 0 |
| Aggregations | 0 | 0 |
| Precedence | 0 | 0 |
| Variable Bound | 0 | 0 |
| Set Partitioning | 78 | 78 |
| Set Packing | 0 | 0 |
| Set Covering | 0 | 0 |
| Cardinality | 0 | 0 |
| Invariant Knapsack | 0 | 0 |
| Equation Knapsack | 0 | 0 |
| Bin Packing | 0 | 0 |
| Knapsack | 0 | 0 |
| Integer Knapsack | 0 | 0 |
| Mixed Binary | 0 | 0 |
| General Linear | 223 | 223 |
| Indicator | 0 | 0 |
Structure
Available nonzero structure and decomposition information. Further information can be found here.

Decomposed structure of original problem (dec-file)

Decomposed structure after trivial presolving (dec-file)
| value | min | median | mean | max | |
|---|---|---|---|---|---|
| Components | |||||
| Constraint % | |||||
| Variable % | |||||
| Score |
Best Known Solution(s)
Find solutions below. Download the archive containing all solutions from the Download page.
## Warning in lapply(df["exactobjval"], as.numeric): NAs introduced by coercion| ID | Objective | Exact | Int. Viol | Cons. Viol | Obj. Viol | Submitter | Date | Description |
|---|---|---|---|---|---|---|---|---|
| 3 | 3382 | 3382 | 0 | 0 | 0 | Edward Rothberg | 2020-09-14 | Found with Gurobi |
| 2 | 3389 | 0 | 0 | 0 | Robert Ashford and Alkis Vazacopoulus | 2019-12-18 | Found using ODH|CPlex | |
| 1 | 3405 | 3405 | 0 | 0 | 0 | - | 2018-10-12 | Solution found during MIPLIB2017 problem selection. |
Similar instances in collection
The following instances are most similar to proteindesign121hz512p19 in the collection. This similarity analysis is based on 100 scaled instance features describing properties of the variables, objective function, bounds, constraints, and right hand sides.
| Instance | Status | Variables | Binaries | Integers | Continuous | Constraints | Nonz. | Submitter | Group | Objective | Tags |
|---|---|---|---|---|---|---|---|---|---|---|---|
| proteindesign121hz512p9 | easy | 159145 | 159054 | 91 | 0 | 301 | 629449 | Gleb Belov | proteindesign | 1473 | benchmark benchmark_suitable set_partitioning general_linear |
| proteindesign121pgb11p9 | easy | 132672 | 132594 | 78 | 0 | 254 | 524690 | Gleb Belov | proteindesign | 1209 | benchmark_suitable set_partitioning general_linear |
| proteindesign122trx11p8 | easy | 127326 | 127248 | 78 | 0 | 254 | 503427 | Gleb Belov | proteindesign | 1747 | benchmark benchmark_suitable set_partitioning general_linear |
| neos-4413714-turia | easy | 190402 | 190201 | 0 | 201 | 2303 | 761756 | Jeff Linderoth | neos-pseudoapplication-67 | 45.3701670199998 | benchmark benchmark_suitable set_partitioning binpacking mixed_binary |
| rvb-sub | hard | 33765 | 33763 | 0 | 2 | 225 | 984143 | S. Weider | – | 16.08499802 | binary set_partitioning knapsack |
Reference
@Inbook{Belov2016,
author="Belov, Gleb
and Stuckey, Peter J.
and Tack, Guido
and Wallace, Mark",
editor="Rueher, Michel",
title="Improved Linearization of Constraint Programming Models",
bookTitle="Principles and Practice of Constraint Programming: 22nd International Conference, CP 2016, Toulouse, France, September 5-9, 2016, Proceedings",
year="2016",
publisher="Springer International Publishing",
pages="49--65",
isbn="978-3-319-44953-1",
doi="10.1007/978-3-319-44953-1_4",
url="http://dx.doi.org/10.1007/978-3-319-44953-1_4"
}Last Update 2024 by Julian Manns
generated with R Markdown
© by Zuse Institute Berlin (ZIB)
Imprint